33 GU Pairs
GU pairs are treated as nearest neighbor stacks, similar to Watson-Crick-Franklin helices, and GU pairs at the ends of helices are not penalized with the same parameter as AU pairs at the ends of helices. In one sequence context, a tandem GU pair with a GU followed by a UG, the nearest neighbor model does not work and two parameters are available, depending on the sequence context (see the html table of parameters). Note also that the motif \(5'\)GG/\(3'\)UU is assigned a ΔG°37 of –0.5 kcal/mol to optimize structure prediction accurracy, whereas it is measured as +0.5 kcal/mol. Parameters for stacks containing GU pairs were calculated separately from those containing AU and GC base pairs only.
33.1 Examples
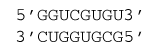
ΔG°37 = ΔG°37 intermolecular initiation + ΔG°37(GC followed by GU, followed by UG, followed by CG) + ΔG°37(CG followed by GU) + ΔG°37(GU followed by UG) + ΔG°37(UG followed by GC) + ΔG°37(GC followed by UG)
ΔG°37 = 4.1 kcal/mol – 4.12 kcal/mol - 1.3 kcal/mol + 0.7 kcal/mol - 1.3 kcal/mol - 2.2 kcal/mol
ΔG°37 = -4.1 kcal/mol
33.2 Parameter Tables
The tables of parameters are available as plain text for free energy change (including Watson-Crick-Franklin pairs).
