23 Dangling Ends
Dangling ends are nucleotides that stack on the ends of helices. In secondary structures, they occur in multibranch and exterior loops. They occur as either \(5'\) dangling ends (an unpaired nucleotide \(5'\) to the helix end) or \(3'\) dangling ends (an unpaired nucleotide \(3'\) to the helix end). Note that if a helix end is extended on both the \(5'\) and \(3'\) strands, then a terminal mismatch exists (not the sum of \(5'\) and \(3'\) dangling ends).
23.1 Examples
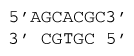
ΔG°37 = ΔG°37(Watson-Crick-Franklin Helix) + ΔG°37(\(3'\) dangling C adajacent to GC) + ΔG°37(\(5'\) dangling A adjacent CG)
ΔG°37 = -6.8 kcal/mol - 0.2 kcal/mol - 0.8 kcal/mol
ΔG°37 = -7.8 kcal/mol
ΔH° = ΔH°(Watson-Crick-Franklin Helix) + ΔH°(\(3'\) dangling C adajacent to GC) + ΔH°(\(5'\) dangling A adjacent CG)
ΔH° = -33.1 kcal/mol -0.2 kcal/mol -4.6 kcal/mol
ΔH° = -37.9 kcal/mol
Note that this example contains both a \(5'\) and a \(3'\) dangling end (at opposite ends of the duplex).
23.2 Parameter Tables
The tables of parameters are available as plain text for free energy change and enthalpy change.
